Project
Background
Although there are thousands of antibiotics that have been found and synthesized in the world, some pathogenic bacteria are multiresistant to antibiotics and extremely hard to be eliminated from the human body. For this reason, scientists have been focus on improving present antibiotics and finding other potential antimicrobials. Nevertheless, the process would be extremely time-consuming and relative costly.
To tackle this serious issue, we came up with a potential solution - expressing suicide genes in antibiotic-resistant bacteria to interrupt their biofunctions. Eventually, we can not only eliminate the bacteria efficiently but cure infected patients without bringing any side effects and cell damage.
First, we thought of a way to deliver genes into bacteria is to form a pore by DNA origami. Later, we found that in previous research ” Synthetic lipid membrane channels formed by designed DNA Nanostructures”, the scientists created transmembrane channels in lipid bilayers using DNA-based nanostructures[1]. Also in this essay, it mentioned that scaffolded DNA origami were used to create a stem that penetrates and spans a lipid membrane, and a barrel-shaped cap that adheres to the membrane via 26 cholesterol. Inspired by this paper, we referred to the DNA origami concept and designed an injector called NanoNeedle. NanoNeedle is able to identify specific targets through aptamers and perforate into bacteria.
Aptamers, the single-stranded DNA molecules consisting of oligonucleotides, are the most essential elements we used for the main function of NanoNeedle. Aptamers are powerful and interesting molecules that will bind to specific targets; therefore, we attached the designed aptamers to the weak structures of NanoNeedle for recognizing components of bacterial cell wall. It includes the endotoxin lipopolysaccharide ( LPS ) and outer membrane proteins. As a result, NanoNeedle will be able to deliver the target insert, suicide genes, into the target bacteria to cause death of bacteria and eventually cure diseases.
Origami design
Our idea is to create a nano-structure. It has the ability to identify the specific pathogenic bacteria, and deliver drugs or plasmid into it.
Like the syringe, where the plunger expels drugs from the tube into the man's body through the needle, we create a structure which enables us to deliver plasmid in it, and expel them into the target bacteria.
The NanoNeedle is constituted of 55 DNA double helices. 42 of the double helices have 300 base pairs each that are used to constitute strong structures and weak structures. The other 13 double helices which are much shorter are used to constitute the cover of the "syringe". Looking at the NanoNeedle from the “cover” perspective, three strong structures and three weak structures are arranged in a stagger pattern to form the body of the NanoNeedle (i.e. strong, weak, strong, weak, strong, weak structures in either a clockwise or a counterclockwise direction).
-
Body
This part of structure is to carry plasmid and stretch to the part that penetrates the cell wall. To meet our goal, we consider to design the shape of the body to be a hollow hexagonal prism.
In order to construct the body part that carries plasmid, we need to evaluate the size first. We regard DNA as a cylinder with radius equal to 1.1nm, and the distance of each base pair is equal to 0.34nm. The plasmid we choose is about 3000 base pairs long. Now we can evaluate the volume of the plasmid:
(1.1)2π * 0.34 * 3000 = 3877.354(nm)3--------(1)
To construct a proper body, we also take our target ---- E. coli into consideration. The cell of E. coli is typically rod-shaped, and is about 2.0 μm long and 0.25–1.0 μm in diameter, with a cell volume of 0.6–0.7 μm3. In order not to kill E. coli when NanoNeedle penetrates into the cell, the structure cannot be too big. We design the hole of the cavity of the body is about 16 cross-sections of DNA, that is
(1.1)2π*16=60.821(nm)2--------(2)
Divide equation (1) by equation (2), we can evaluate the minimal length of the body is approximately 63.750nm, that is about 190 base pairs.
As a whole, the body part is a hollow hexagonal prism with inner diameter of about 4.400nm, outer diameter of about 18.000nm, and the length is about 63.750nm.
In addition, to prevent plasmid from escaping from the body, we design some short double helices to seal the plasmid in the structure. There are 13 shorter double helices to constitute the cover on the top. DNA origami should form the enclosed top with mesh structure.
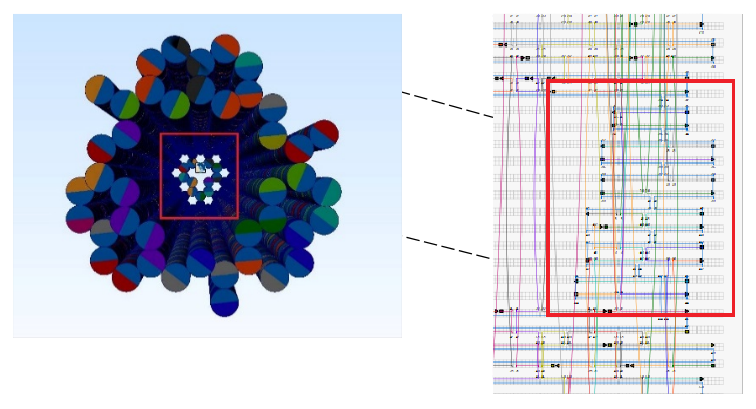
13 shorter double helices constitute the cover of the NanoNeedle -
Strong structures
The syringe uses a needle to stab through the skin. NanoNeedle also has a similar structure, which we call strong structures. Each strong structure is constituted of ten DNA double helices. For the first 84 base pairs, each double helix sticks with each other by using staples, but unlike the body structure, there is no crossover between strong structures and weak structures. While weak structures stick to the cell wall, strong structures will penetrate and span the cell wall and membrane due to their solid structure.
-
Weak structures
Weak structures are soft enough to bend while strong structures stab through the bacteria. Aptamers are equipped on weak structures for several uses.
-
Aptamers
There are three uses for aptamers. One is identifying and binding with cell walls. Due to the difference of affinity between aptamers, the weak structures can bend outward, thus, NanoNeedle can insert plasmid into the bacteria. Second is closing the opening.The last one is binding with the plasmid.
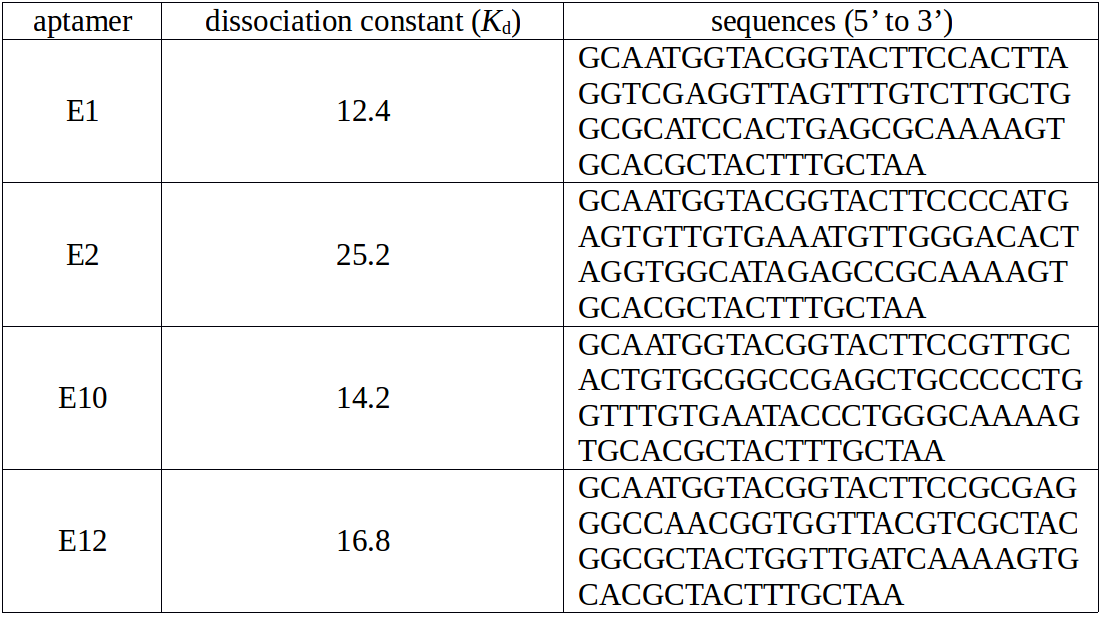
NanoNeedle targets at E. coli our experiment design mentioned before, thus we search out aptamers only binding to E. coli in previous research of DNA aptamers against E. coli using by SELEX [2]. In this research, scientists finally isolated four ssDNA aptamers (E1, E2, E10 and E12) showing high affinity and selectivity to E. coli. The possible targets of these aptamers are lipopolysaccharide (LPS), membrane proteins and flagella on outer membrane of E. coli. The information of these aptamers is in the following table.
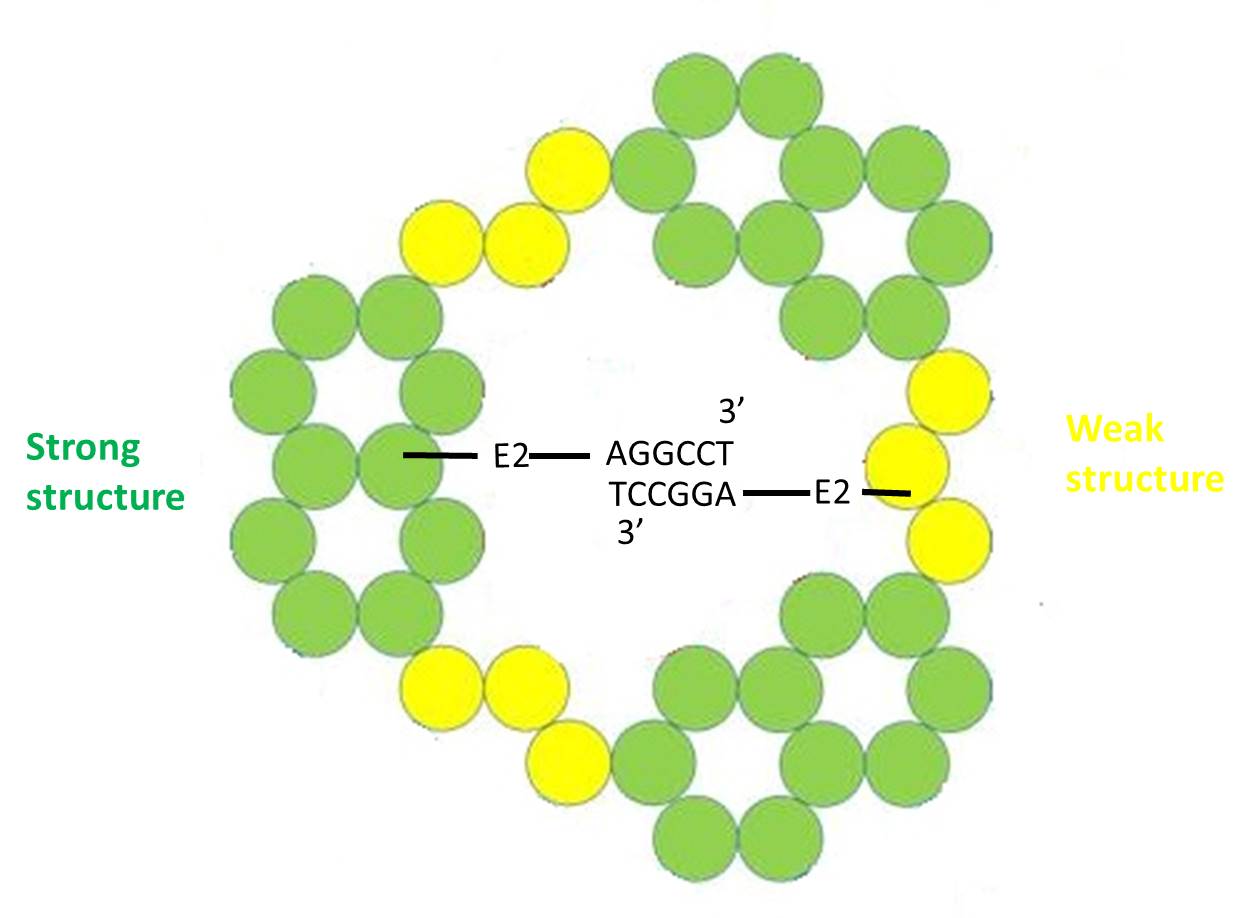
We chose three ssDNA aptamers, E2, E10 and E1, to bind from bottom to middle of weak structure with increasing-affinity.
Except for E1, 3’ end of E2 and E10 bind to additional sequences which we design. Each designed aptamer will perform its own function.
-
E2
E2 binds to AGGCCT(5’ to 3’) complementary to AGGCCT binding to 3’ end of staple of opposite strong structure. However we cannot ensure base-pairings are all between opposite, they may form between two sides. There are 3 base-pairings are between opposite in optimal condition but at least one will able to close the bottom of NanoNeedle to prevent plasmid moving out of the structure.
-
E10
E10 binds to CTGCA complementary to sticky ends of pSB1C3-BBa_I746909 digested by restriction enzyme PstI as showed in following picture.
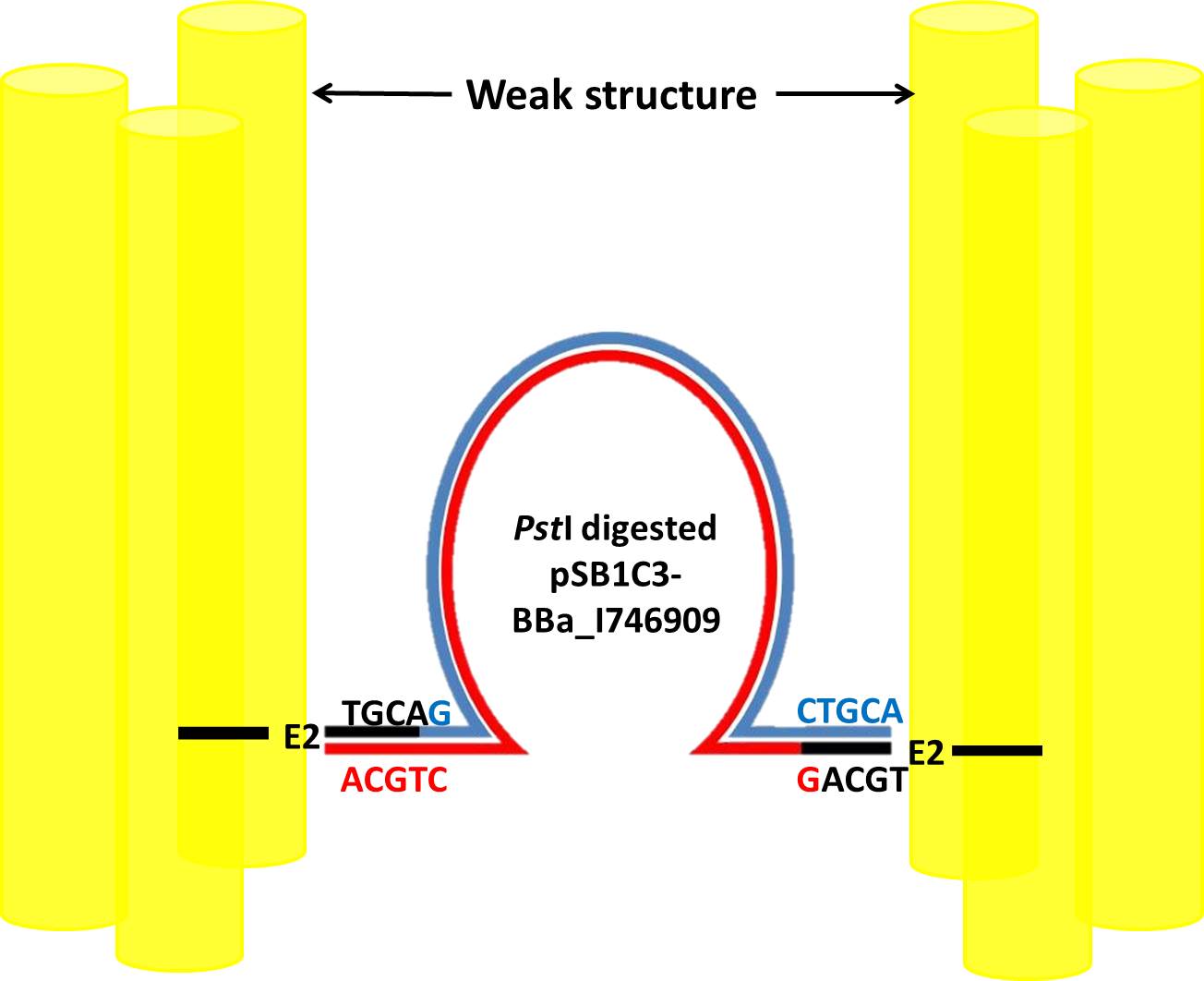
The restriction map of the pSB1C3-BBa_I746909 and the recognition site of PstI are showed in following pictures.
This base-paring can make sure plasmid stays inside of NanoNeedle until E10 binds toE. coli.

The restriction map of pSB1C3-BBa_I746909
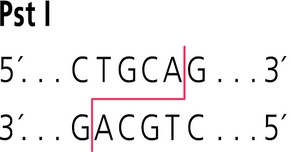 The recognition site of restriction enzyme PstI
The recognition site of restriction enzyme PstI
-
E2
-
E1
E1 with greatest affinity binds to middle of weak structure to make sure that NanoNeedle span through the cell membrane of E. coli.
Through our origami design, NanoNeedle will penetrate E. coli through three steps. When NanoNeedle meets E. coli, E2 first binds to components on outer membrane and the weak structure starts to bend. Because affinity of E10 is higher than E2, E10 secondly binds to components bound by E2 before and then releases the plasmid.
Finally E1 binds to the components and the strong structure of NanoNeedle penetrates through the cell membrane to ensure plasmid enter cytoplasm of the E. coli.
Using CaDNAno2, we can construct the whole NanoNeedle. The structure (i.e. strong structure, weak structure, body) is composed of two templates, p8064 and M13mp18, and staples generated by the build-in function. NanoNeedle should fold in the shape of hexagonal column and form the enclosed top with mesh structure.
 Structure design using CaDNAno2
Structure design using CaDNAno2
Experiment Design
In our project NanoNeedle will perform three functions:
First is to specifically recognize the target antibiotic-resistant bacteria. Second is to penetrate the cell wall and membrane, and finally deliver plasmid into bacteria.
In our experiment design, we aim to check NanoNeedle can function well and also establish an experimental model for future application.
First we choose E. coli, a well-known model organism, as our target bacteria.Second we choose a plasmid carries the Green fluorescent protein (GFP) gene to bind to aptamers inside NanoNeedle because in E. coli system the bright green fluorescence exhibited by GFP can be a marker for us to know whether plasmid is successfully delivered into E. coli. If E. coli incubated with NanoNeedle shows GFP signal when exposed to blue light in flow cytrometry, we can verify that the plasmid is well expressed and NanoNeedle successfully performs its functions on E. coli.
Here we design 5 parts of experiments.
-
Construction a plasmid with the GFP gene
We choose a plasmid pSB1C3-BBa_I746909 to be connected to aptamers in NanoNeedle. This plasmid, carrying superfolder the GFP gene driven by T7-promoter, can be transformed into E. coli and successfully encode superfolder GFP, a robustly folded version of GFP.
The initial amount of plasmids is not enough for further experiments, thus we conduct transformation, mini-preparation, midi-preparation and extraction of plasmid to acquire sufficient plasmids.
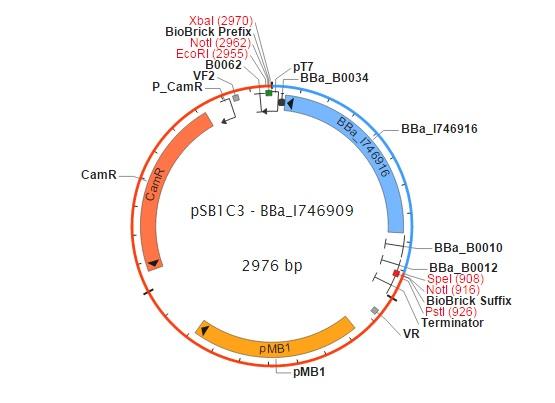
figure 1 The map of pSB1C3-BBa_I746909 (synthesized by Cambridge group in iGEM)
-
DNA folding reaction
Since the concentration of magnesium ion affects the structure of DNA origami significantly, we first conduct the folding reaction of scaffolds and staples and electrophoresis analysis to determine the best concentration of magnesium ion.
Then we use restriction enzyme PstI to digest pSB1C3-BBa_I746909 and digested plasmid is able to stay inside NanoNeedle through binding to complimentary sequence at aptamers. Finally we perform complete folding reactions and electrophoresis analysis to test whether our DNA origami is folded.
-
TEM imaging
The 3D structure of NanoNeedle is then confirmed by transmission electron microscopy (TEM).
-
Functional test
After TEM imaging to confirm NanoNeedle is folded into correct structure, we need to test its three functions. Here we incubate target bacteria E. coli with NanoNeedle for few hours to test whether E. coli shows obvious GFP signal measured by flow cytometry.
-
Cell culture cytotoxicity
In the future we expect to kill bacteria by delivering NanoNeedle into patients suffered from bacterial infection; therefore we need to confirm that NanoNeedle will not injure human cells by MTT assay.
MTT assay is a colorimetric assay for assessing cell metabolic activity. MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a yellow tetrazole, is reduced to insoluble purple formazan by Succinate dehydrogenase (SDH) in mitochondria of living cells. Dimethyl sulfoxide (DMSO) is added to dissolve formazan into a colored solution. The absorbance of this colored solution can be quantified by measuring at a certain wavelength (usually between 500 and 600 nm) and estimated how many cells are alive.
We choose HT29 Cell Line (originates from Human Caucasian colon adenocarcinoma) to be incubated with NanoNeedle and then use ELISA reader to measure absorbance of 595nm for testing the cytotoxicity.
_______________________________
[1]Langecker, M., Arnaut, V., Martin T.G., et al.(2012). Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936.
[2]Kim, Y.S., Song, M.Y., Jurng, J., et al.(2013).Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Anal. Biochem 436, 22–28


